It is thought that some epigenetic information can be altered by extrinsic or environmental factors, such as diet and early life experiences, and such changes could be perpetuated long after these exposures and potentially to future generations. Such epigenetic programming may contribute to our risk of developing disease in later life.
The discovery that gene transcription is essential for patterning much of the epigenome of the egg, including establishing imprints, may help explain how epigenetic marks in the egg could be modified by extrinsic factors such as altered maternal diet. We are currently deploying our single-cell profiling methods (Kelsey et al. 2017) to explore whether DNA methylation and gene expression in oocytes are modified by maternal diet, and whether such changes persist in embryos. We are also investigating whether some procedures associated with assisted reproduction technologies, for example hormone stimulation for egg retrieval, modify the epigenome of the egg or embryo.
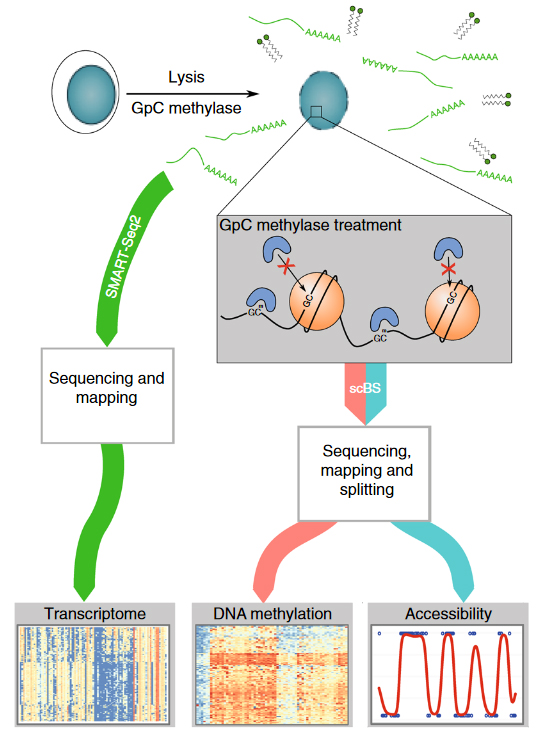 Combined mapping of chromatin, methylation and expression in single oocytes.
Combined mapping of chromatin, methylation and expression in single oocytes.
Overview of the scNMT-seq method. Single-cells are lysed and accessible DNA is labelled using GpC methyltransferase. RNA is then separated and sequenced using Smart-seq2, whilst DNA undergoes scBS-seq library preparation and sequencing. Methylation and chromatin accessibility data are separated bioinformatically. We applying this method to identify expression and methylation alterations that may be elicited by altered maternal diet. From Clark et al. (2018) scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Communications http://dx.doi.org/10.1038/s41467-018-03149-4.