How DNA is packaged in cells and the use of biochemical switches in the genome are key aspects of the epigenetic control of gene activity. We are interested in understanding how epigenetic processes are established during human development and during the differentiation of stem cells to form various cell types. This is important for understanding health and for finding ways to use stem cells in regenerative biology.
Importance of epigenetic programming in development and regenerative biology
Pluripotent epiblast cells in the human embryo, highlighted in green in the figure above, undergo a series of lineage decisions during the first two weeks of development, which are essential for the successful implantation, growth and patterning of the conceptus. During this period, cells must retain information about their identity including past fate decisions, and prepare them for further differentiation events. Similar processes also control epiblast-derived pluripotent stem cells that are cultured in the laboratory.
Epigenetic mechanisms create opportunities in development for controlling cell identity. Combinations of chromatin proteins and their associated histone marks localise to regulatory sequences, and these elements interact with transcription factors to execute cellular programmes. Epigenetic processes can modulate these responses to endow cells with developmental plasticity associated with the sustained capacity for multi-lineage differentiation, and to instruct future cell fate decisions.
We investigate epigenetic programming in developing human embryos and human pluripotent stem cells including embryo-like models. This research is important because understanding how epigenetic processes are established and influence embryo development sheds light on how cell lineages are correctly defined and ultimately lead to a healthy pregnancy. Our research may help to improve rates of embryo development and could identify new molecular markers that indicate successful development, leading to crucial benefits to reproductive health. Our discoveries into the regulators of pluripotent cells and how they specialise also makes important contributions to regenerative biology.
Please see our Human Embryo Development webpage and the Human Developmental Biology Initiative web site for more information.
Controlling developmental gene activity
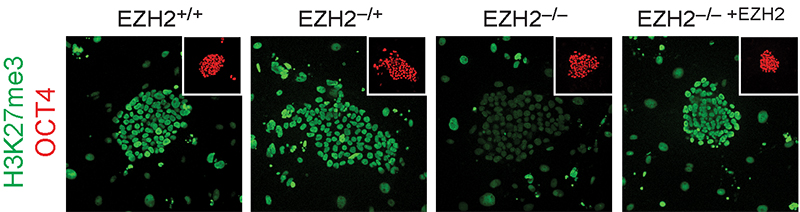
Focusing on the regulation of early lineage-decisions, we characterised the first EZH2-deficient human pluripotent stem cells and found there is a broad conservation of Polycomb-group protein function in controlling cell-fate decisions and transcriptional programs during early human development doi.org/10.1016/j.celrep.2016.11.032 (press release) (see figure below). We also uncovered unexpected human-specific differences that result in a more severe self-renewal and proliferation phenotype than that of Polycomb-deficient mouse pluripotent stem cells.
Expanding our mechanistic understanding of how developmental genes are controlled, we demonstrated that epigenetically-primed regions are spatially connected in human pluripotent stem cells, developed new computational tools to study this, and reported how these epigenetic features are established as cells undergo state transitions doi.org/10.1038/s41467-021-22201-4; doi.org/10.7554/eLife.21926 (press release).
Leveraging this knowledge, we worked with a team of international collaborators to discover that naïve-state human pluripotent stem cells are not epigenetically unrestricted as previously thought, but instead possess chromatin-based mechanisms that oppose the induction of alternative cell fates including trophoblast doi.org/10.1038/s41556-022-00932-w (press release). Functional studies in stem cells and integrated embryo models led us to propose that these chromatin regulators are pivotal mediators of the earliest lineage specification events (see figure below).
Defining cell states and the origins of embryo lineages
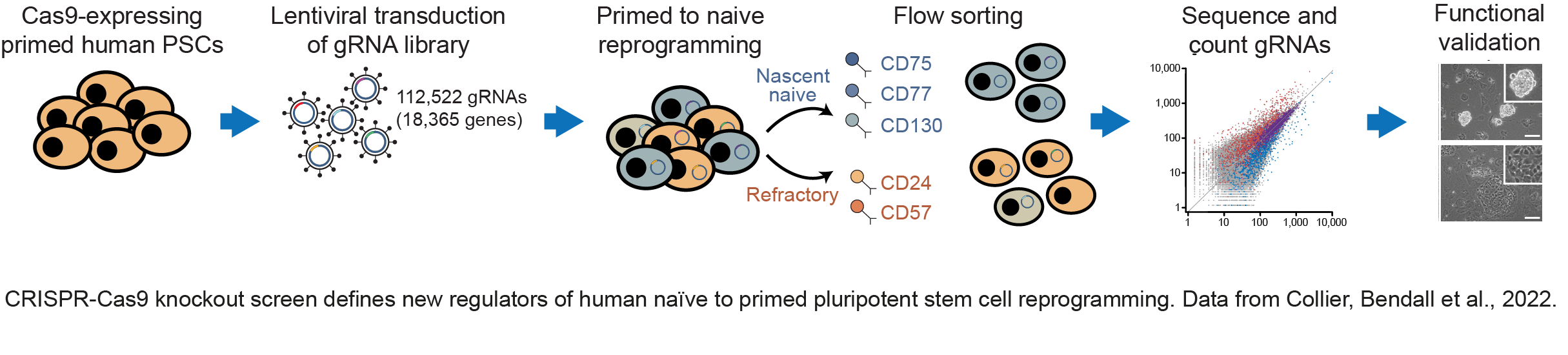
We have made important contributions to understanding how distinct epigenetic states are established in pluripotent cells, and how those states control the entry and exit of pluripotency. We have identified cell-surface markers that accurately define mouse and human pluripotent cell types doi.org/10.1016/j.stem.2017.02.014; doi.org/10.1016/j.stemcr.2020.03.017; doi.org/10.1016/j.devcel.2012.01.005) (press release). Taking advantage of these markers, we have completed a genome-wide genetic screen to identify factors that control the transitions between human pluripotent states doi.org/10.1126/sciadv.abk0013 (press release) (see figure below). The findings allowed us to propose new mechanistic leads in cell state control. The work also has important practical implications as we were able to design rational approaches to improve on current methodologies to induce changes in pluripotent state; a finding that we anticipate will open up new ways to exploit the full potential of human pluripotent cells.
Lastly, together with Maria Rostovskaya, Simon Andrews and Wolf Reik, we discovered that human pluripotent stem cells transiently gain the ability to form amnion-like cells as they transition from naïve to primed pluripotency doi.org/10.1016/j.stem.2022.03.014 (see figure below). This has striking parallels to human epiblast development in embryos. As amnion cells have beneficial properties that can promote wound healing, this discovery could lead to new ways to generate valuable cell-based therapies.
Defining new connections between pluripotency and genome organisation
An important goal is to better understand what triggers the epigenome to be remodelled at specific stages of development. On these lines, we found that pluripotency transcription factors provide a direct connection between cell-state and chromatin organisation through modulation of heterochromatin regions in mouse pluripotent stem cells doi.org/10.1101/gad.275685.115 (press release) (see figure below). These changes are controlled by modulating the transcription of major satellite repeat sequences, which are abundant in heterochromatin, and this alters the behaviour and properties of heterochromatin doi.org/10.1038/s41467-022-31198-3. This mode of regulation is important because changes in heterochromatin organisation affect centromere function and chromosome stability in pluripotent cells and during cell reprogramming.
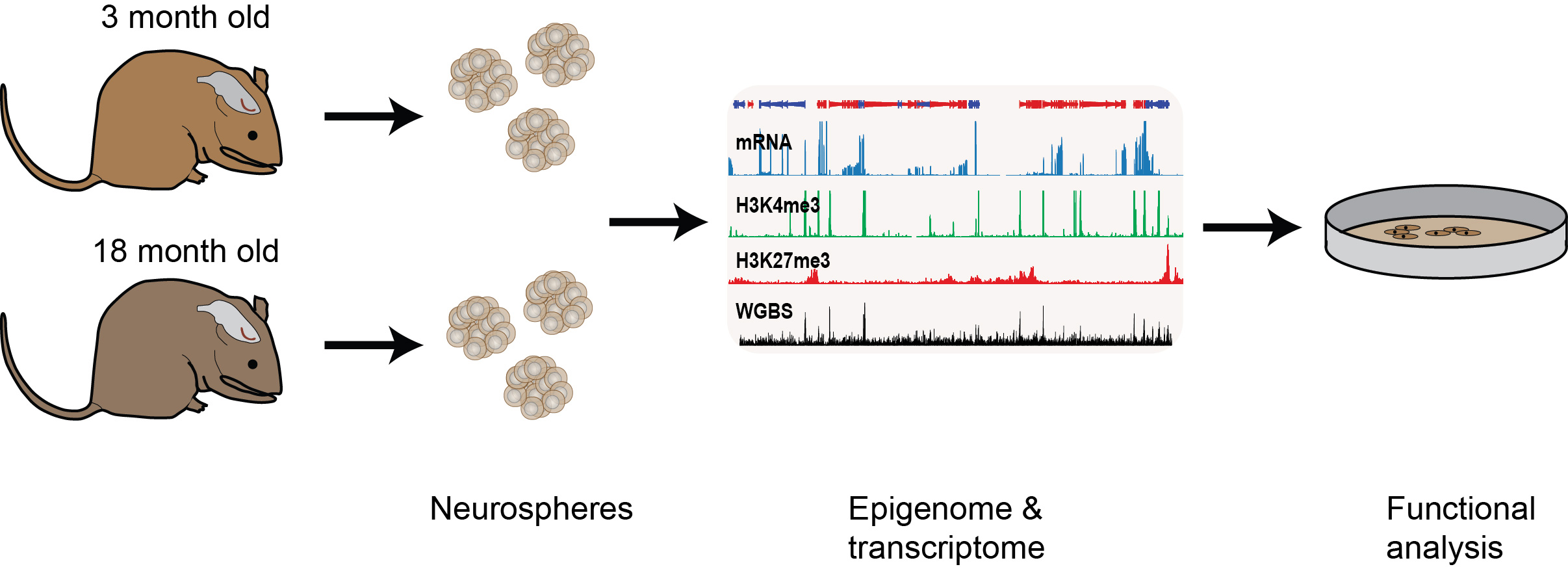
Epigenetic changes associated with the decline of stem cell function upon ageing
We are also interested in understanding how epigenetic information changes over the life course of an individual. For example, adult neural stem progenitor cell (NSPC) function declines with ageing but the underlying molecular causes are largely unknown. We are examining the intrinsic changes that occur upon NSPC ageing through genome-wide transcriptional, histone methylation and DNA methylation analyses of NSPCs derived from the subventricular zone of adult (3 months old) and aged (18 months old) mice. We found that the genome-wide profiles were largely unchanged upon ageing, however, within this large data set, we also identified significant transcriptional and epigenetic differences at several hundred loci doi.org/10.1111/acel.12745 (press release). We are now following up on these results to see if we can uncover new regulators of age-associated neurogenic decline and neural stem cell function