The idea that genome change can be regulated in response to the environment has major implications. It is generally thought that genome changes leading to adaptation occur through random mutation followed by selection, however, we are accumulating evidence that cells can focus genome changes on certain genomic regions in response to the environment, accelerating adaptation. We have demonstrated that this occurs in the ribosomal DNA, and more recently shown that protein coding genes can be similarly subject to regulated, stimulated genetic change. This provides a rapid path by which cells can adapt to thrive in challenging environments
Understanding how adaptive genetic changes occur is critical: from the acquisition of drug resistance by tumour cells to the emergence of non-productive mutants in industrial fermentation processes, adaptive genetic changes impact our health and industrial productivity. The assumption that adaptive genetic change is random suggests that it is unavoidable, but we propose that cells use active, regulated pathways to adapt, and that these pathways can themselves form drug targets to prevent the adaptation process.
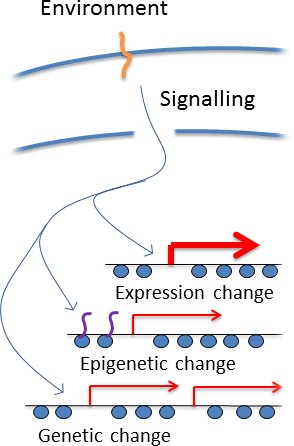 How could cells regulate genetic change? This is an old problem, and the biggest issue with Lamarkian inheritance. It is though that regulated genetic change requires a cell to 'know' the effect of a particular change in advance to decide what to do. In genomes containing millions or billions of individual basepairs, this is implausible. However, this argument ignores the wealth of information
How could cells regulate genetic change? This is an old problem, and the biggest issue with Lamarkian inheritance. It is though that regulated genetic change requires a cell to 'know' the effect of a particular change in advance to decide what to do. In genomes containing millions or billions of individual basepairs, this is implausible. However, this argument ignores the wealth of information
about the function of particular genomic regions that is embedded in their gene expression patterns, which are highly evolved to handle changing environments.
All cells have signalling pathways that sesnse particular environments and invoke the induction of particular genes under particular circumstances, which immediately identifies the genomic regions containing those genes as being important in the current environment (see right). It has log been understood that such pathways bring about epigenetic changes, and we simply suggest that cells couuld also target mutations such as copy number variation to the same loci. So a cell does not 'know' what genetic changes to make in a particular environment, but it appliles the available information to take an educated guess at which genomic regions are particularly good for mutation. This is not a perfect solution but would be good enough to substantially accelerate adaptation.
Model systems for studying genome change
Protein coding loci
5-10% of genes in the human genome occur more than once in the haploid genome (ie: are multi-copy), and the same is true in most other eukaryotic genomes. We study the dynamics of copy number change in protein coding genes in yeast to determine the extent to which copy number change is regulated.
The clearest example to date is the CUP1 gene. This encodes a protein responsible for copper resistance in yeast, and we have recently demonstrated that copy number variation of CUP1 is driven by transcription that is in turn stimulated in response to the environment (Hull et al. PLoS Biology, 2017). This gives rise to a spectrum of novel CUP1 alleles as predicted above, from which new variants with higher copper resistance are selected. This process is controlled by a network of signalling pathways and chromatin remodelling activities, and by blocking these pathways we can inhibit the adaptation of yeast to copper. This is the best evidence yet that it may be possible to generally control the adaptive process.
The mechanisms we have identified are not restricted to the CUP1 gene, and we predict that ~7% of yeaast genes have the potential to undergo copy number variation stimulated in response to a diverse range of environments.
Ribosomal DNA
We also study the yeast ribosomal DNA as a model of regulated copy number variation. This region contains ~150 tandem copies and ribosomal DNA copy number is carefully maintained. Cells with too few copies of the ribosomal DNA cannot produce sufficient ribosomes for maximum growth, and rapidly amplify the ribosomal DNA copy number back to normal levels.
This remains by far the clearest example of copy number variation as a regulated process in any organism. Non-protein coding RNA produced from the ribosomal DNA is involved in regulating copy number, and we have found that proteins which interact with these RNAs are important for copy number regulation (see below).
Surprisingly, we have found that copy number amplification occurs by a highly unusual mechanism which does not require the well-understood homologous recombination machinery. Instead, amplification proceeds through a poorly understood mechanism previously known for causing occasional pathogenic chromosomal rearrangements.
Furthermore, we have shown that ribosomal DNA copy number is controlled by a specific signalling pathway, TOR, which responds to nutrient availability, allowing cells to specifically optimise their genome for current environmental conditions.
 Ribosomal DNA recombination is tightly linked to ageing in yeast, and many of the mutations we have found that alter ribosomal DNA recombination also impact lifespan. We are very interested in understanding how regulated recombination processes can change lifespan, as some of these mechanisms are likely to be highly conserved amongst eukaryotes.
Ribosomal DNA recombination is tightly linked to ageing in yeast, and many of the mutations we have found that alter ribosomal DNA recombination also impact lifespan. We are very interested in understanding how regulated recombination processes can change lifespan, as some of these mechanisms are likely to be highly conserved amongst eukaryotes.